Magnetoencephalography
Contents
Magnetoencephalography#
Support for Magnetoencephalography (MEG) was developed as a BIDS Extension Proposal. Please see Citing BIDS on how to appropriately credit this extension when referring to it in the context of the academic literature.
The following example MEG datasets have been formatted using this specification and can be used for practical guidance when curating a new dataset.
Further datasets are available from the BIDS examples repository.
MEG recording data#
Unprocessed MEG data MUST be stored in the native file format of the MEG instrument with which the data was collected. With the MEG specification of BIDS, we wish to promote the adoption of good practices in the management of scientific data. Hence, the emphasis is not to impose a new, generic data format for the modality, but rather to standardize the way data is stored in repositories. Further, there is currently no widely accepted standard file format for MEG, but major software applications, including free and open-source solutions for MEG data analysis, provide readers of such raw files.
Some software readers may skip important metadata that is specific to MEG system manufacturers. It is therefore RECOMMENDED that users provide additional meta information extracted from the manufacturer raw data files in a sidecar JSON file. This allows for easy searching and indexing of key metadata elements without the need to parse files in proprietary data format. Other relevant files MAY be included alongside the MEG data; examples are provided below.
This template is for MEG data of any kind, including but not limited to
task-based, resting-state, and noise recordings.
If multiple Tasks were performed within a single Run,
the task description can be set to task-multitask.
The *_meg.json file SHOULD contain details on the Tasks.
Some manufacturers’ data storage conventions use directories which contain data
files of various nature: for example, CTF’s .ds format, or BTi/4D’s data directory.
Yet other manufacturers split their files once they exceed a certain size
limit.
For example Neuromag/Elekta/Megin, which can produce several files
for a single recording.
Both some_file.fif and some_file-1.fif would belong to a single recording.
In BIDS, the split entity is RECOMMENDED to deal
with split files.
If there are multiple parts of a recording and the optional scans.tsv is provided,
remember to list all files separately in scans.tsv and that the entries for the
acq_time column in scans.tsv MUST all be identical, as described in
Scans file.
Another manufacturer-specific detail pertains to the KIT/Yokogawa/Ricoh system,
which saves the MEG sensor coil positions in a separate file with two possible filename extensions (.sqd, .mrk).
For these files, the markers suffix MUST be used.
For example: sub-01_task-nback_markers.sqd
Please refer to Appendix VI for general information on how to deal with such manufacturer specifics and to see more examples.
The proc-<label> entity is analogous to the
rec-<label> entity for MRI,
and denotes a variant of a file that was a result of particular processing performed on the device.
This is useful for files produced in particular by Elekta’s MaxFilter
(for example, sss, tsss, trans, quat, mc),
which some installations impose to be run on raw data prior to analysis.
Such processing steps are needed for example because of active shielding software corrections
that have to be performed to before the MEG data can actually be exploited.
Recording EEG simultaneously with MEG#
Note that if EEG is recorded with a separate amplifier,
it SHOULD be stored separately under a new /eeg data type
(see the EEG specification).
If however EEG is recorded simultaneously with the same MEG system,
it MAY be stored under the /meg data type.
In that case, it SHOULD have the same sampling frequency as MEG (see SamplingFrequency field below).
Furthermore, the EEG sensor coordinates SHOULD be specified using MEG-specific coordinate
systems (see coordinates section below and Appendix VIII).
Sidecar JSON (*_meg.json)#
Generic fields MUST be present:
SHOULD be present: For consistency between studies and institutions, we encourage users to extract the values of these fields from the actual raw data. Whenever possible, please avoid using ad-hoc wording.
Specific MEG fields MUST be present:
SHOULD be present:
Specific EEG fields (if recorded with MEG, see Recording EEG simultaneously with MEG SHOULD be present:
Example:
{
"InstitutionName": "Stanford University",
"InstitutionAddress": "450 Serra Mall, Stanford, CA 94305-2004, USA",
"Manufacturer": "CTF",
"ManufacturersModelName": "CTF-275",
"DeviceSerialNumber": "11035",
"SoftwareVersions": "Acq 5.4.2-linux-20070507",
"PowerLineFrequency": 60,
"SamplingFrequency": 2400,
"MEGChannelCount": 270,
"MEGREFChannelCount": 26,
"EEGChannelCount": 0,
"EOGChannelCount": 2,
"ECGChannelCount": 1,
"EMGChannelCount": 0,
"DewarPosition": "upright",
"SoftwareFilters": {
"SpatialCompensation": {"GradientOrder": "3rd"}
},
"RecordingDuration": 600,
"RecordingType": "continuous",
"EpochLength": 0,
"TaskName": "rest",
"ContinuousHeadLocalization": true,
"HeadCoilFrequency": [1470,1530,1590],
"DigitizedLandmarks": true,
"DigitizedHeadPoints": true
}
Note that the date and time information SHOULD be stored in the Study key file
(scans.tsv), see Scans file.
Date time information MUST be expressed as indicated in Units
Channels description (*_channels.tsv)#
This file is RECOMMENDED as it provides easily searchable information across BIDS datasets.
For example for general curation, response to queries, or for batch analysis.
To avoid confusion, the channels SHOULD be listed in the order they appear in the MEG data file.
Any number of additional columns MAY be added to provide additional information about the channels.
Missing values MUST be indicated with "n/a".
The columns of the channels description table stored in *_channels.tsv are:
MUST be present in this specific order:
SHOULD be present:
Example:
name type units description sampling_frequency low_cutoff high_cutoff notch software_filters status
UDIO001 TRIG V analogue trigger 1200 0.1 300 0 n/a good
MLC11 MEGGRADAXIAL T sensor 1st-order grad 1200 0 n/a 50 SSS bad
Restricted keyword list for field type.
Note that upper-case is REQUIRED:
Keyword |
Description |
|---|---|
MEGMAG |
MEG magnetometer |
MEGGRADAXIAL |
MEG axial gradiometer |
MEGGRADPLANAR |
MEG planargradiometer |
MEGREFMAG |
MEG reference magnetometer |
MEGREFGRADAXIAL |
MEG reference axial gradiometer |
MEGREFGRADPLANAR |
MEG reference planar gradiometer |
MEGOTHER |
Any other type of MEG sensor |
EEG |
Electrode channel |
ECOG |
Electrode channel |
SEEG |
Electrode channel |
DBS |
Electrode channel |
VEOG |
Vertical EOG (electrooculogram) |
HEOG |
Horizontal EOG |
EOG |
Generic EOG channel |
ECG |
ElectroCardioGram (heart) |
EMG |
ElectroMyoGram (muscle) |
TRIG |
System Triggers |
AUDIO |
Audio signal |
PD |
Photodiode |
EYEGAZE |
Eye Tracker gaze |
PUPIL |
Eye Tracker pupil diameter |
MISC |
Miscellaneous |
SYSCLOCK |
System time showing elapsed time since trial started |
ADC |
Analog to Digital input |
DAC |
Digital to Analog output |
HLU |
Measured position of head and head coils |
FITERR |
Fit error signal from each head localization coil |
OTHER |
Any other type of channel |
Example of free text for field description:
stimulus, response, vertical EOG, horizontal EOG, skin conductance, sats, intracranial, eyetracker
Example:
name type units description
VEOG VEOG V vertical EOG
FDI EMG V left first dorsal interosseous
UDIO001 TRIG V analog trigger signal
UADC001 AUDIO V envelope of audio signal presented to participant
Coordinate System JSON (*_coordsystem.json)#
OPTIONAL. A JSON document specifying the coordinate system(s) used for the MEG, EEG, head localization coils, and anatomical landmarks.
MEG and EEG sensors:
Head localization coils:
Digitized head points:
Anatomical MRI:
Anatomical landmarks:
It is also RECOMMENDED that the MRI voxel coordinates of the actual anatomical
landmarks for co-registration of MEG with structural MRI are stored in the
AnatomicalLandmarkCoordinates field in the JSON sidecar of the corresponding
T1w MRI anatomical data of the subject seen in the MEG session
(see Anatomy Imaging Data).
For example: "sub-01/ses-mri/anat/sub-01_ses-mri_acq-mprage_T1w.json"
In principle, these locations are those of absolute anatomical markers. However,
the marking of NAS, LPA and RPA is more ambiguous than that of for example, AC and PC.
This may result in some variability in their 3-D digitization from session to
session, even for the same participant. The solution would be to use only one
T1w file and populate the AnatomicalLandmarkCoordinates field with
session-specific labels for example, “NAS-session1”: [127,213,139],”NAS-session2”:
[123,220,142].
Fiducials information:
For more information on the definition of anatomical landmarks, please visit: http://www.fieldtriptoolbox.org/faq/how_are_the_lpa_and_rpa_points_defined
For more information on typical coordinate systems for MEG-MRI coregistration: http://www.fieldtriptoolbox.org/faq/how_are_the_different_head_and_mri_coordinate_systems_defined, or: http://neuroimage.usc.edu/brainstorm/CoordinateSystems
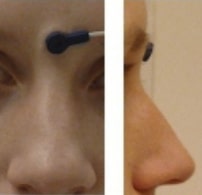
Landmark photos (*_photo.jpg)#
Photos of the anatomical landmarks and/or head localization coils
(*_photo.jpg)
Photos of the anatomical landmarks and/or head localization coils on the subject’s head are RECOMMENDED. If the coils are not placed at the location of actual anatomical landmarks, these latter may be marked with a piece of felt-tip taped to the skin. Please note that the photos may need to be cropped or blurred to conceal identifying features prior to sharing, depending on the terms of the consent given by the participant.
The acq-<label> entity can be used to indicate acquisition of different photos of
the same face (or other body part in different angles to show, for example, the
location of the nasion (NAS) as opposed to the right periauricular point (RPA)).
Example of the NAS fiducial placed between the eyebrows, rather than at the
actual anatomical nasion: sub-0001_ses-001_acq-NAS_photo.jpg
Head shape and electrode description (*_headshape.<ext>)#
This file is RECOMMENDED.
The 3-D locations of points that describe the head shape and/or EEG
electrode locations can be digitized and stored in separate files. The
acq-<label> entity can be used when more than one type of digitization in done for
a session, for example when the head points are in a separate file from the EEG
locations. These files are stored in the specific format of the 3-D digitizer’s
manufacturer (see Appendix VI).
Example:
Note that the *_headshape file(s) is shared by all the runs and tasks in a
session. If the subject needs to be taken out of the scanner and the head-shape
has to be updated, then for MEG it could be considered to be a new session.
Empty-room MEG recordings#
Empty-room MEG recordings capture the environmental and recording system’s
noise.
In the context of BIDS it is RECOMMENDED to perform an empty-room recording for
each experimental session.
It is RECOMMENDED to store the empty-room recording inside a subject directory
named sub-emptyroom.
The label for the task-<label> entity in the empty-room recording SHOULD be
set to noise.
If a session-<label> entity is present, its label SHOULD be the date of the
empty-room recording in the format YYYYMMDD, that is ses-YYYYMMDD.
The scans.tsv file containing the date and time of the acquisition SHOULD
also be included.
The rationale is that this naming scheme will allow users to easily retrieve the
empty-room recording that best matches a particular experimental session, based
on date and time of the recording.
It should be possible to query empty-room recordings just like usual subject
recordings, hence all metadata sidecar files (such as the channels.tsv) file
SHOULD be present as well.
Example:

