Intracranial Electroencephalography
Contents
Intracranial Electroencephalography#
Support Intracranial Electroencephalography (iEEG) was developed as a BIDS Extension Proposal. Please see Citing BIDS on how to appropriately credit this extension when referring to it in the context of the academic literature.
Several example iEEG datasets have been formatted using this specification and can be used for practical guidance when curating a new dataset.
iEEG recording data#
The iEEG community uses a variety of formats for storing raw data, and there is no single standard that all researchers agree on. For BIDS, iEEG data MUST be stored in one of the following formats:
Format |
Extension(s) |
Description |
|---|---|---|
|
Each recording consists of a |
|
|
Each recording consists of a |
|
|
The format used by the MATLAB toolbox EEGLAB. Each recording consists of a |
|
|
Each recording consists of a single |
|
|
Each recording consists of a |
It is RECOMMENDED to use the European data format, or the BrainVision data format. It is furthermore discouraged to use the other accepted formats over these RECOMMENDED formats, particularly because there are conversion scripts available in most commonly used programming languages to convert data into the RECOMMENDED formats.
Future versions of BIDS may extend this list of supported file formats. File formats for future consideration MUST have open access documentation, MUST have open source implementation for both reading and writing in at least two programming languages and SHOULD be widely supported in multiple software packages. Other formats that may be considered in the future should have a clear added advantage over the existing formats and should have wide adoption in the BIDS community.
The data format in which the data was originally stored is especially valuable
in case conversion elicits the loss of crucial metadata specific to
manufacturers and specific iEEG systems. We also encourage users to provide
additional meta information extracted from the manufacturer-specific data files
in the sidecar JSON file. Other relevant files MAY be included alongside the
original iEEG data in the /sourcedata directory.
Note the RecordingType, which depends on whether the data stream on disk is interrupted or not. Continuous data is by definition 1 segment without interruption. Epoched data consists of multiple segments that all have the same length (for example, corresponding to trials) and that have gaps in between. Discontinuous data consists of multiple segments of different length, for example due to a pause in the acquisition.
Terminology: Electrodes vs. Channels#
For proper documentation of iEEG recording metadata it is important to understand the difference between electrode and channel: an iEEG electrode is placed on or in the brain, whereas a channel is the combination of the analog differential amplifier and analog-to-digital converter that result in a potential (voltage) difference that is stored in the iEEG dataset. We employ the following short definitions:
Electrode = A single point of contact between the acquisition system and the recording site (for example, scalp, neural tissue, …). Multiple electrodes can be organized as arrays, grids, leads, strips, probes, shafts, caps (for EEG), and so forth.
Channel = A single analog-to-digital converter in the recording system that regularly samples the value of a transducer, which results in the signal being represented as a time series in the digitized data. This can be connected to two electrodes (to measure the potential difference between them), a magnetic field or magnetic gradient sensor, temperature sensor, accelerometer, and so forth.
Although the reference and ground electrodes are often referred to as channels, they are in most common iEEG systems not recorded by themselves. Therefore they are not represented as channels in the data. The type of referencing for all channels and optionally the location of the reference electrode and the location of the ground electrode MAY be specified.
Sidecar JSON (*_ieeg.json)#
For consistency between studies and institutions, we encourage users to extract the values of metadata fields from the actual raw data. Whenever possible, please avoid using ad hoc wording.
Generic fields MUST be present:
Note that the TaskName field does not have to be a “behavioral task” that subjects perform, but can reflect some information about the conditions present when the data was acquired (for example, "rest", "sleep", or "seizure").
SHOULD be present: For consistency between studies and institutions, we encourage users to extract the values of these fields from the actual raw data. Whenever possible, please avoid using ad hoc wording.
Specific iEEG fields MUST be present:
Specific iEEG fields SHOULD be present:
Specific iEEG fields MAY be present:
Example:
{
"TaskName":"visual",
"InstitutionName":"Stanford Hospital and Clinics",
"InstitutionAddress":"300 Pasteur Dr, Stanford, CA 94305",
"Manufacturer":"Tucker Davis Technologies",
"ManufacturersModelName":"n/a",
"TaskDescription":"visual gratings and noise patterns",
"Instructions":"look at the dot in the center of the screen and press the button when it changes color",
"iEEGReference":"left mastoid",
"SamplingFrequency":1000,
"PowerLineFrequency":60,
"SoftwareFilters":"n/a",
"HardwareFilters":{"Highpass RC filter": {"Half amplitude cutoff (Hz)": 0.0159, "Roll-off": "6dBOctave"}},
"ElectrodeManufacturer":"AdTech",
"ECOGChannelCount":120,
"SEEGChannelCount":0,
"EEGChannelCount":0,
"EOGChannelCount":0,
"ECGChannelCount":0,
"EMGChannelCount":0,
"MiscChannelCount":0,
"TriggerChannelCount":0,
"RecordingDuration":233.639,
"RecordingType":"continuous",
"iEEGGround":"placed on the right mastoid",
"iEEGPlacementScheme":"right occipital temporal surface",
"ElectricalStimulation":false
}
Note that the date and time information SHOULD be stored in the Study key file
(scans.tsv).
Date time information MUST be expressed as indicated in Units
Channels description (*_channels.tsv)#
A channel represents one time series recorded with the recording system
(for example, there can be a bipolar channel, recorded from two electrodes or contact points on the tissue).
Although this information can often be extracted from the iEEG recording,
listing it in a simple .tsv document makes it easy to browse or search
(for example, searching for recordings with a sampling frequency of >=1000 Hz).
Hence, the channels.tsv file is RECOMMENDED.
Channels SHOULD appear in the table in the same order they do in the iEEG data file.
Any number of additional columns MAY be provided to provide additional information about the channels.
Note that electrode positions SHOULD NOT be added to this file but to *_electrodes.tsv.
The columns of the channels description table stored in *_channels.tsv are:
MUST be present in this specific order:
SHOULD be present:
Example sub-01_channels.tsv:
name type units low_cutoff high_cutoff status status_description
LT01 ECOG uV 300 0.11 good n/a
LT02 ECOG uV 300 0.11 bad broken
H01 SEEG uV 300 0.11 bad line_noise
ECG1 ECG uV n/a 0.11 good n/a
TR1 TRIG n/a n/a n/a good n/a
Restricted keyword list for field type in alphabetic order (shared with the MEG and EEG modality; however, only types that are common in iEEG data are listed here). Note that upper-case is REQUIRED:
Keyword |
Description |
|---|---|
EEG |
Electrode channel from electroencephalogram |
ECOG |
Electrode channel from electrocorticogram (intracranial) |
SEEG |
Electrode channel from stereo-electroencephalogram (intracranial) |
DBS |
Electrode channel from deep brain stimulation electrode (intracranial) |
VEOG |
Vertical EOG (electrooculogram) |
HEOG |
Horizontal EOG |
EOG |
Generic EOG channel if HEOG or VEOG information not available |
ECG |
ElectroCardioGram (heart) |
EMG |
ElectroMyoGram (muscle) |
TRIG |
System Triggers |
AUDIO |
Audio signal |
PD |
Photodiode |
EYEGAZE |
Eye Tracker gaze |
PUPIL |
Eye Tracker pupil diameter |
MISC |
Miscellaneous |
SYSCLOCK |
System time showing elapsed time since trial started |
ADC |
Analog to Digital input |
DAC |
Digital to Analog output |
REF |
Reference channel |
OTHER |
Any other type of channel |
Example of free-form text for field description:
intracranial, stimulus, response, vertical EOG, skin conductance
Electrode description (*_electrodes.tsv)#
File that gives the location, size and other properties of iEEG electrodes. Note
that coordinates are expected in cartesian coordinates according to the
iEEGCoordinateSystem and iEEGCoordinateUnits fields in
*_coordsystem.json. If an *_electrodes.tsv file is specified, a
*_coordsystem.json file MUST be specified as well.
The optional space-<label> entity (*[_space-<label>]_electrodes.tsv) can be used to
indicate the way in which electrode positions are interpreted.
The space <label> MUST be taken from one of the modality specific lists in
Appendix VIII.
For example for iEEG data, the restricted keywords listed under
iEEG Specific Coordinate Systems
are acceptable for <label>.
For examples:
_space-MNI152Lin(electrodes are coregistred and scaled to a specific MNI template)_space-Talairach(electrodes are coregistred and scaled to Talairach space)
When referring to the *_electrodes.tsv file in a certain space as defined
above, the space-<label> of the accompanying *_coordsystem.json MUST
correspond.
For example:
The order of the required columns in the *_electrodes.tsv file MUST be as listed below.
The x, y, and z columns indicate the positions of the center of each electrode in Cartesian coordinates.
Units are specified in space-<label>_coordsystem.json.
MUST be present in this specific order:
SHOULD be present:
MAY be present:
Example:
name x y z size manufacturer
LT01 19 -39 -16 2.3 Integra
LT02 23 -40 -19 2.3 Integra
H01 27 -42 -21 5 AdTech
Coordinate System JSON (*_coordsystem.json)#
This _coordsystem.json file contains the coordinate system in which electrode
positions are expressed. The associated MRI, CT, X-Ray, or operative photo can
also be specified.
General fields:
Fields relating to the iEEG electrode positions:
Recommended 3D coordinate systems#
It is preferred that electrodes are localized in a 3D coordinate system (with respect to a pre- and/or post-operative anatomical MRI or CT scans or in a standard space as specified in BIDS Appendix VIII about preferred names of coordinate systems, such as ACPC).
Allowed 2D coordinate systems#
If electrodes are localized in 2D space (only x and y are specified and z is "n/a"),
then the positions in this file MUST correspond to the locations expressed
in pixels on the photo/drawing/rendering of the electrodes on the brain.
In this case, iEEGCoordinateSystem MUST be defined as "Pixels",
and iEEGCoordinateUnits MUST be defined as "pixels"
(note the difference in capitalization).
Furthermore, the coordinates MUST be (row,column) pairs,
with (0,0) corresponding to the upper left pixel and (N,0) corresponding to the lower left pixel.
Multiple coordinate systems#
If electrode positions are known in multiple coordinate systems (for example, MRI, CT
and MNI), these spaces can be distinguished by the optional space-<label>
field, see the *_electrodes.tsv-section
for more information.
Note that the space-<label> fields must correspond
between *_electrodes.tsv and *_coordsystem.json if they refer to the same
data.
Example:
{
"IntendedFor": "/sub-01/ses-01/anat/sub-01_T1w.nii.gz",
"iEEGCoordinateSystem": "ACPC",
"iEEGCoordinateUnits": "mm",
"iEEGCoordinateSystemDescription": "Coordinate system with the origin at anterior commissure (AC), negative y-axis going through the posterior commissure (PC), z-axis going to a mid-hemisperic point which lies superior to the AC-PC line, x-axis going to the right",
"iEEGCoordinateProcessingDescription": "surface_projection",
"iEEGCoordinateProcessingReference": "Hermes et al., 2010 JNeuroMeth"
}
Photos of the electrode positions (*_photo.jpg)#
These can include photos of the electrodes on the brain surface, photos of anatomical features or landmarks (such as sulcal structure), and fiducials. Photos can also include an X-ray picture, a flatbed scan of a schematic drawing made during surgery, or screenshots of a brain rendering with electrode positions. The photos may need to be cropped and/or blurred to conceal identifying features or entirely omitted prior to sharing, depending on obtained consent.
If there are photos of the electrodes, the acq-<label> entity should be specified
with:
*_photo.jpgin case of an operative photo*_acq-xray#_photo.jpgin case of an x-ray picture*_acq-drawing#_photo.jpgin case of a drawing or sketch of electrode placements*_acq-render#_photo.jpgin case of a rendering
The ses-<label> entity may be used to specify when the photo was taken.
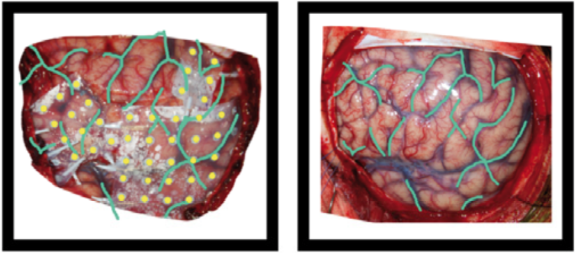
Example of the operative photo of ECoG electrodes (here is an annotated example in which electrodes and vasculature are marked, taken from Hermes et al., JNeuroMeth 2010).
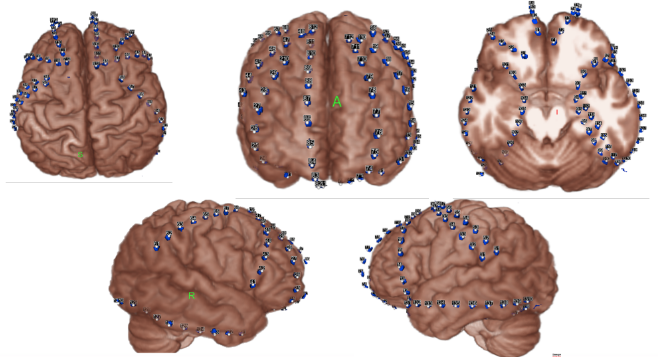
Below is an example of a volume rendering of the cortical surface with a superimposed subdural electrode implantation. This map is often provided by the
EEG technician and provided to the epileptologists (for example, see Burneo JG et al. 2014. doi:10.1016/j.clineuro.2014.03.020).
sub-0002_ses-01_acq-render_photo.jpg
Electrical stimulation#
In case of electrical stimulation of brain tissue by passing current through the
iEEG electrodes, and the electrical stimulation has an event structure (on-off,
onset, duration), the _events.tsv file can contain the electrical stimulation
parameters in addition to other events. Note that these can be intermixed with
other task events. Electrical stimulation parameters can be described in columns
called electrical_stimulation_<label>, with labels chosen by the researcher and
optionally defined in more detail in an accompanying _events.json file (as
per the main BIDS spec). Functions for complex stimulation patterns can, similar
as when a video is presented, be stored in a directory in the /stimuli/ directory.
For example: /stimuli/electrical_stimulation_functions/biphasic.tsv
Example:
onset duration trial_type electrical_stimulation_type electrical_stimulation_site electrical_stimulation_current
1.2 0.001 electrical_stimulation biphasic LT01-LT02 0.005
1.3 0.001 electrical_stimulation biphasic LT01-LT02 0.005
2.2 0.001 electrical_stimulation biphasic LT02-LT03 0.005
4.2 1 electrical_stimulation complex LT02-LT03 n/a
15.2 3 auditory_stimulus n/a n/a n/a

